The Volume Library
The intention of the library is to provide volume datasets for
scientists involved with volume visualization and
rendering. Commercial use is prohibited and no warranty whatsoever is
expressed.
The datasets contain regular volume data mainly coming from CT or MRI scanners. The data is stored in the PVM format which contains information about the grid size, bit depth, and the cell spacing of a dataset. Optionally it may also contain a dataset description, courtesy information, the type of the scanner and a comment. This information and the raw data can be extracted easily using the PVM tools distributed with the V^3 volume rendering package available at my sourceforge page. Just checkout the V^3 package, type "install.sh" in a Linux shell and use the pvminfo resp. pvm2raw utility in the tools folder to extract the dataset information resp. the raw data.
Bits [Aspect] Checksum | Additional Comments | |||||
 |  | Download (7608kb) | 8 bit 0.974019/0.974655/1.48125 0C16BF8F | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | ||
 | Download (3152kb) | 8 bit 0.583649/0.583649/1.00586 C4A176F2 | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | |||
 |  | Download (1844kb) | 8 bit 311D75D8 | Jason Bryan is the developer and maintainer of VolSuite. Feel free to email him with questions or comments about the software: jbryan at osc dot edu | ||
 |  | Download (20436kb) | 8 bit 0.9766/0.9766/1.25 BD3512BD | |||
 |  | Download (196kb) | 8 bit 1/0.75/1 31FA79A8 | http://www.nas.nasa.gov/Research/Datasets/datasets.html | ||
 |  | non-linear quantized version Download (22384kb) | 8 bit 0.585938/0.585938/1 4D9DEE6A | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | Siemens Somatom Plus 4 16.Oct.1999 Thanks to Bernd Tomandl | |
 | 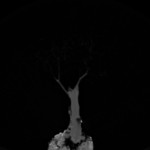 | linear quantized version Download (10356kb) | 8 bit 0.585938/0.585938/1 23D5981D | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | Siemens Somatom Plus 4 16.Oct.1999 Thanks to Bernd Tomandl | |
 | 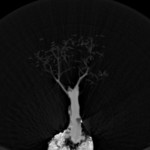 | Download (24508kb) | 16 bit 0.585938/0.585938/1 AF985501 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | Siemens Somatom Plus 4 16.Oct.1999 Thanks to Bernd Tomandl | |
 | 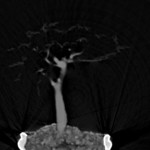 | non-linear quantized version Download (17000kb) | 8 bit 0.402344/0.402344/1 865F7616 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | Siemens Somatom Plus 4 16.Oct.1999 Thanks to Bernd Tomandl | |
 |  | linear quantized version Download (9736kb) | 8 bit 0.402344/0.402344/1 AF0BDAC8 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | Siemens Somatom Plus 4 16.Oct.1999 Thanks to Bernd Tomandl | |
 |  | Download (22416kb) | 16 bit 0.402344/0.402344/1 A84386C0 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | Siemens Somatom Plus 4 16.Oct.1999 Thanks to Bernd Tomandl | |
 |  | non-linear quantized version Download (12824kb) | 8 bit 0.402344/0.402344/1 8487BBB5 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | Siemens Somatom Plus 4 16.Oct.1999 Thanks to Bernd Tomandl | Bonsai from IEEE Visualization '00 Conference |
 |  | linear quantized version Download (7944kb) | 8 bit 0.402344/0.402344/1 48FA5E11 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | Siemens Somatom Plus 4 16.Oct.1999 Thanks to Bernd Tomandl | Bonsai from IEEE Visualization '00 Conference |
 |  | Download (18428kb) | 16 bit 0.402344/0.402344/1 415A4E0A | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | Siemens Somatom Plus 4 16.Oct.1999 Thanks to Bernd Tomandl | Bonsai from IEEE Visualization '00 Conference |
 |  | Download (16kb) | 8 bit 07F7C3E4 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | Test dataset containing a solid box | |
 |  | Download (7444kb) | 16 bit 7E53970E | University of Utah, Salt Lake City http://www.cs.utah.edu/~bgooch/BruceBrain.html | July 13th, 2001 | The scans were taken as part of an arcane research project by a buddy of mine named Jim Lee. Jim also provided the Matlab scripts you can use to view the MRI data. I am making the data freely available with the following caveats: 1: If you publish a scientific paper with an image created using my MRI data you will mention that the image was created using the "Bruce Gooch's Brain" data sets. 2: If you create a really cool image of my brain you will email me a copy of it. I will post all such images on this site. Thank You, and happy computing. Bruce Gooch June 12, 2001 |
 |  | Download (24kb) | 8 bit 102CDE9F | http://www.avs.com | ||
 |  | Download (64956kb) | 16 bit 0.337891/0.337891/0.5 F9A197D4 | High Performance Computing and Communications, National Library of Medicine, USA mailto:yoo@nlm.nih.gov | 28.Jan.2000 | This is a CT scan of the terracotta Stanford Bunny. Three hundred sixty-one files comprise the bunny. The scale of the voxel data is 0.337891 mm x 0.337891 mm x 0.5mm in the x-, y-, and z- dimensions respectively. The greyscale units are Hounsfield units, denoting electron-density of the subject. The data is raw 512x512 slices, unsigned, 12 bit data stored as 16bit (2-byte) pixels. The scan was completed 28 January 2000. Many many thanks to Geoff Rubin who helped me to scan the data, Sandy Napel who coordinated the scan and helped to process the data, and Marc Levoy who graciously provided the subject. Geoff and Sandy are with Stanford Radiology, and Marc is with Stanford Computer Science. Dataset contact info: Terry S. Yoo High Performance Computing and Communications yoo@nlm.nih.gov National Library of Medicine National Institutes of Health |
 | Download (23992kb) | 8 bit 0.779724/0.779724/4.96599 AAAA125E | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | |||
 |  | Download (7104kb) | 8 bit AB7D36C1 | http://radiology.uiowa.edu/downloads | ||
 |  | Download (5616kb) | 16 bit 0.95/1/1.8878 5728224B | Computer Graphics Laboratory, Stanford University, USA http://graphics.stanford.edy/~levoy Provided courtesy of North Carolina Memorial Hospital | General Electric CT Scanner | |
 |  | Download (7316kb) | 8 bit 8D6A4540 | http://radiology.uiowa.edu/downloads | ||
 |  | Download (9692kb) | 8 bit 0.214844/0.214844/0.5 39317391 | http://www.neuroradiologie.med.uni-erlangen.de | EBE CTA 1 dataset | |
 |  | Download (16480kb) | 16 bit 0.435547/0.435547/2 02C1D792 | http://www9.cs.fau.de | The cadaver head is famous for the scanning artifacts originating at the teeth | |
 |  | Download (19864kb) | 16 bit 0.78125/0.390625/1 AEB61B35 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de | The Carp is a seasonal delicacy in Frankonia, Germany | |
 |  | Download (836kb) | 8 bit 743C02CB | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | ||
 | Download (2756kb) | 8 bit 1.27992/1.27992/4.88636 0564C110 | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | |||
 | Download (3636kb) | 8 bit 1.35906/1.35906/2.94915 503F9522 | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | |||
 |  | Download (12kb) | 8 bit 8724C5A0 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de | Test dataset containing several crossed rods, balls and platters Rods are placed like coordinate axes to verify correct orientation | |
 |  | Download (716kb) | 16 bit 4977B2FE | http://www.cs.unc.edu/~fillard | B0 = lores MRI scan (no diffusion weighting) B1 = diffusion weighted volume #1 with dir=(1,0,1) B2 = diffusion weighted volume #2 with dir=(-1,0,1) B3 = diffusion weighted volume #3 with dir=(0,1,1) B4 = diffusion weighted volume #4 with dir=(0,1,-1) B5 = diffusion weighted volume #5 with dir=(1,1,0) B6 = diffusion weighted volume #6 with dir=(-1,1,0) MD = mean diffusion FA = fractional anisotropy | |
 |  | Download (580kb) | 16 bit 424F4A19 | http://www.cs.unc.edu/~fillard | B0 = lores MRI scan (no diffusion weighting) B1 = diffusion weighted volume #1 with dir=(1,0,1) B2 = diffusion weighted volume #2 with dir=(-1,0,1) B3 = diffusion weighted volume #3 with dir=(0,1,1) B4 = diffusion weighted volume #4 with dir=(0,1,-1) B5 = diffusion weighted volume #5 with dir=(1,1,0) B6 = diffusion weighted volume #6 with dir=(-1,1,0) MD = mean diffusion FA = fractional anisotropy | |
 | 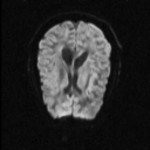 | Download (580kb) | 16 bit AAB49B39 | http://www.cs.unc.edu/~fillard | B0 = lores MRI scan (no diffusion weighting) B1 = diffusion weighted volume #1 with dir=(1,0,1) B2 = diffusion weighted volume #2 with dir=(-1,0,1) B3 = diffusion weighted volume #3 with dir=(0,1,1) B4 = diffusion weighted volume #4 with dir=(0,1,-1) B5 = diffusion weighted volume #5 with dir=(1,1,0) B6 = diffusion weighted volume #6 with dir=(-1,1,0) MD = mean diffusion FA = fractional anisotropy | |
 | 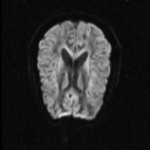 | Download (580kb) | 16 bit 488DAC6A | http://www.cs.unc.edu/~fillard | B0 = lores MRI scan (no diffusion weighting) B1 = diffusion weighted volume #1 with dir=(1,0,1) B2 = diffusion weighted volume #2 with dir=(-1,0,1) B3 = diffusion weighted volume #3 with dir=(0,1,1) B4 = diffusion weighted volume #4 with dir=(0,1,-1) B5 = diffusion weighted volume #5 with dir=(1,1,0) B6 = diffusion weighted volume #6 with dir=(-1,1,0) MD = mean diffusion FA = fractional anisotropy | |
 |  | Download (576kb) | 16 bit EFA22F4E | http://www.cs.unc.edu/~fillard | B0 = lores MRI scan (no diffusion weighting) B1 = diffusion weighted volume #1 with dir=(1,0,1) B2 = diffusion weighted volume #2 with dir=(-1,0,1) B3 = diffusion weighted volume #3 with dir=(0,1,1) B4 = diffusion weighted volume #4 with dir=(0,1,-1) B5 = diffusion weighted volume #5 with dir=(1,1,0) B6 = diffusion weighted volume #6 with dir=(-1,1,0) MD = mean diffusion FA = fractional anisotropy | |
 |  | Download (584kb) | 16 bit 11743FB5 | http://www.cs.unc.edu/~fillard | B0 = lores MRI scan (no diffusion weighting) B1 = diffusion weighted volume #1 with dir=(1,0,1) B2 = diffusion weighted volume #2 with dir=(-1,0,1) B3 = diffusion weighted volume #3 with dir=(0,1,1) B4 = diffusion weighted volume #4 with dir=(0,1,-1) B5 = diffusion weighted volume #5 with dir=(1,1,0) B6 = diffusion weighted volume #6 with dir=(-1,1,0) MD = mean diffusion FA = fractional anisotropy | |
 |  | Download (584kb) | 16 bit FF48D5B6 | http://www.cs.unc.edu/~fillard | B0 = lores MRI scan (no diffusion weighting) B1 = diffusion weighted volume #1 with dir=(1,0,1) B2 = diffusion weighted volume #2 with dir=(-1,0,1) B3 = diffusion weighted volume #3 with dir=(0,1,1) B4 = diffusion weighted volume #4 with dir=(0,1,-1) B5 = diffusion weighted volume #5 with dir=(1,1,0) B6 = diffusion weighted volume #6 with dir=(-1,1,0) MD = mean diffusion FA = fractional anisotropy | |
 |  | Download (648kb) | 8 bit E62446AC | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | B0 = lores MRI scan (no diffusion weighting) B1 = diffusion weighted volume #1 with dir=(1,0,1) B2 = diffusion weighted volume #2 with dir=(-1,0,1) B3 = diffusion weighted volume #3 with dir=(0,1,1) B4 = diffusion weighted volume #4 with dir=(0,1,-1) B5 = diffusion weighted volume #5 with dir=(1,1,0) B6 = diffusion weighted volume #6 with dir=(-1,1,0) MD = mean diffusion FA = fractional anisotropy | |
 | 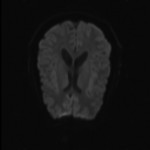 | Download (364kb) | 8 bit 7F461F36 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | B0 = lores MRI scan (no diffusion weighting) B1 = diffusion weighted volume #1 with dir=(1,0,1) B2 = diffusion weighted volume #2 with dir=(-1,0,1) B3 = diffusion weighted volume #3 with dir=(0,1,1) B4 = diffusion weighted volume #4 with dir=(0,1,-1) B5 = diffusion weighted volume #5 with dir=(1,1,0) B6 = diffusion weighted volume #6 with dir=(-1,1,0) MD = mean diffusion FA = fractional anisotropy | |
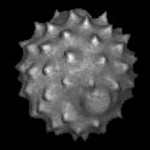 | 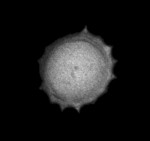 | Download (568kb) | 8 bit 745EFA2F | Computer Science Institute, University of Freiburg, Germany | ||
 |  | Download (2900kb) | 8 bit E786CB95 | http://www.ge.com | GE Industrial Scanner | Two cylinders of an engine block |
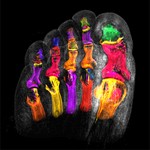 | 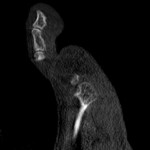 | Download (4592kb) | 8 bit 4FAD56F0 | http://www.philips.de/forschung | Philips CT Scanner | |
 |  | Download (1652kb) | 8 bit 0.5/0.5/1 B2E8D2C2 | http://www-itg.lbl.gov | This is the second frog used in the Whole Frog Project. Three sets of data were produced with the MRI scans. frog2ci.hips seemed to be the best, and only this set of data was processed using HIPS filters to balance the contrast on the images. This same frog was later sliced and a better data set is produced that showed more detail of the frog. frog2ci.hips -- is a spin echo data set with 50 slices, inplane resolution of 0.5mm, slice thickness and separation of 1.0mm, TE = 33ms. The individual slices 2-45 are in the tiff directory. For information about the HIPS file format see http://www-itg.lbl.gov/ITG.hm.pg.docs/image-proc/CCS-ECL/README.txt This data is copyrighted by Lawrence Berkeley National Laboratory. | |
 |  | Download (12kb) | 8 bit 282902A6 | The higher the density value, the lower the presence of air | ||
 |  | Download (196kb) | 8 bit 4A792245 | a hydrogen atom, residing in a strong magnetic field. | ||
 |  | Download (5912kb) | 16 bit 4/4/3 4462EA24 | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger The Weather Research and Forecasting (WRF) Model simulation data of Hurricane Isabel was kindly provided by Bill Kuo, Wei Wang, Cindy Bruyere, Tim Scheitlin, and Don Middleton of the U.S. National Center for Atmospheric Research (NCAR) and the U.S. National Science Foundation (NSF) at this location: http://www.vets.ucar.edu/vg/isabeldata/ | ||
 |  | Download (5772kb) | 16 bit 4/4/3 5039AFFF | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger The Weather Research and Forecasting (WRF) Model simulation data of Hurricane Isabel was kindly provided by Bill Kuo, Wei Wang, Cindy Bruyere, Tim Scheitlin, and Don Middleton of the U.S. National Center for Atmospheric Research (NCAR) and the U.S. National Science Foundation (NSF) at this location: http://www.vets.ucar.edu/vg/isabeldata/ | ||
 | Download (32548kb) | 16 bit 4/4/3 C9695DDD | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger The Weather Research and Forecasting (WRF) Model simulation data of Hurricane Isabel was kindly provided by Bill Kuo, Wei Wang, Cindy Bruyere, Tim Scheitlin, and Don Middleton of the U.S. National Center for Atmospheric Research (NCAR) and the U.S. National Science Foundation (NSF) at this location: http://www.vets.ucar.edu/vg/isabeldata/ | |||
 |  | Download (8384kb) | 8 bit 1.0909/1.0909/0.989583 B23A8009 | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | ||
 | Download (3260kb) | 8 bit 0.466919/0.466919/0.61899 3732AC40 | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | |||
 | 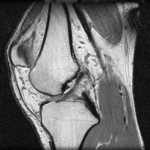 | Download (26040kb) | 16 bit 0.25/0.25/1.5 21D925AD | http://splweb.bwh.harvard.edu | From the Transfer Function Bakeoff at IEEE Visualization '00 Conference Original source: http://visual.nlm.nih.gov | |
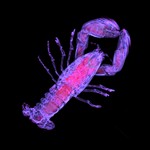 | 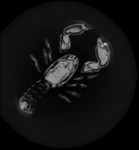 | Download (1908kb) | 8 bit 1/1/1.4 CFCD4D44 | http://www.volvis.org | The lobster is contained in a block of resin | |
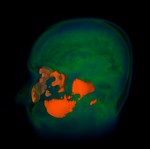 |  | Download (13412kb) | 8 bit 1/1/0.8 3396D681 | http://www9.cs.fau.de | ||
 |  | Download (7420kb) | 16 bit 19FB5371 | Montreal Neurological Institute, McGill University http://www.bic.mni.mcgill.ca/cgi/brainweb | BrainWeb: Simulated Brain Database As the interest in the computer-aided, quantitative analysis of medical image data is growing, the need for the validation of such techniques is also increasing. Unfortunately, there exists no `ground truth' or gold standard for the analysis of in vivo acquired data. The BrainWeb pages provide a solution to the validation problem, in the form of a Simulated Brain Database (SBD). The SBD contains a set of realistic MRI data volumes produced by an MRI simulator. These data can be used by the neuroimaging community to evaluate the performance of various image analysis methods in a setting where the truth is known. PD = MRI Proton Density T1 = MRI T1 measurement T2 = MRI T2 measurement | |
 |  | Download (6656kb) | 16 bit 3AB8A09B | Montreal Neurological Institute, McGill University http://www.bic.mni.mcgill.ca/cgi/brainweb | BrainWeb: Simulated Brain Database As the interest in the computer-aided, quantitative analysis of medical image data is growing, the need for the validation of such techniques is also increasing. Unfortunately, there exists no `ground truth' or gold standard for the analysis of in vivo acquired data. The BrainWeb pages provide a solution to the validation problem, in the form of a Simulated Brain Database (SBD). The SBD contains a set of realistic MRI data volumes produced by an MRI simulator. These data can be used by the neuroimaging community to evaluate the performance of various image analysis methods in a setting where the truth is known. PD = MRI Proton Density T1 = MRI T1 measurement T2 = MRI T2 measurement | |
 |  | Download (7648kb) | 16 bit DEFC2412 | Montreal Neurological Institute, McGill University http://www.bic.mni.mcgill.ca/cgi/brainweb | BrainWeb: Simulated Brain Database As the interest in the computer-aided, quantitative analysis of medical image data is growing, the need for the validation of such techniques is also increasing. Unfortunately, there exists no `ground truth' or gold standard for the analysis of in vivo acquired data. The BrainWeb pages provide a solution to the validation problem, in the form of a Simulated Brain Database (SBD). The SBD contains a set of realistic MRI data volumes produced by an MRI simulator. These data can be used by the neuroimaging community to evaluate the performance of various image analysis methods in a setting where the truth is known. PD = MRI Proton Density T1 = MRI T1 measurement T2 = MRI T2 measurement | |
 |  | Download (7788kb) | 16 bit 1/1/1.5 EA3A8842 | Siemens Magnetom | ||
 |  | Download (540kb) | 8 bit 55513E1F | http://www.loni.ucla.edu/Research/Atlases/ | ||
 |  | Download (2160kb) | 8 bit E0DAD1AC | http://www.loni.ucla.edu/Research/Atlases/ | ||
 |  | Download (1724kb) | 8 bit 2D7656C6 | http://www.loni.ucla.edu/Research/Atlases/ | ||
 |  | Download (3340kb) | RGB 100FE261 | http://www.loni.ucla.edu/Research/Atlases/ | ||
 |  | Download (68kb) | 8 bit 42A8A6EF | http://www.volvis.org | a high potential protein molecule. | Neghip = Negative high potential protein molecule |
 |  | Download (2480kb) | 8 bit 0.390625/0.390625/1 3CA3D462 | Information and Computing Sciences Division, Lawrence Berkeley Laboratory, USA http://www-itg.lbl.gov | These images and data were produced at Lawrence Berkeley Laboratory by Bill Johnston and Wing Nip of the Information and Computing Sciences Division. This work is supported by the U. S. Department of Energy, Energy Research Division, Office of the Scientific Computing Staff under contract DE-AC03-76SF00098. This data is provided for the purpose of scientific research and collaboration with Lawrence Berkeley Laboratory. Any commercial use of these images and data require prior agreement with Lawrence Berkeley Laboratory. The images and data are Copyright Lawrence Berkeley Laboratory. For further information contact Bill Johnston (510-486-5014, wejohnston@lbl.gov). | |
 |  | Download (1060kb) | 8 bit 0.70166/0.701752/0.885938 DCF08EE7 | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | ||
 |  | Download (16516kb) | 16 bit 0.371094/0.371094/1 E0EF8A00 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de | Siemens Volume Zoom Somatom Plus 4 | To prevent scanning artifacts the piggy bank contains chocolate coins |
 |  | Download (32356kb) | 8 bit B05EFB08 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de | ||
 | Download (6500kb) | 16 bit 1.25/1.25/1.85 92EA86AD | ||||
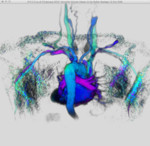 | 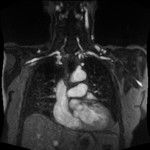 | Download (4152kb) | 16 bit 1.25/1.25/1.85 29A40E5E | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | ||
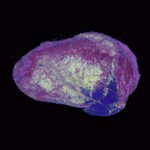 | 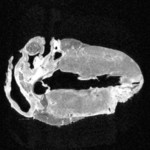 | Download (18772kb) | 8 bit FC68B74E | http://wwwcivm.mc.duke.edu | From the Transfer Function Bakeoff at IEEE Visualization '00 Conference Original source: http://visual.nlm.nih.gov | |
 |  | Download (68kb) | 8 bit DF219C08 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | Test dataset containing the euclidean distance to the origin | |
 |  | Download (1596kb) | 8 bit 4817C587 | Computer Graphics Group, University of Erlangen, Germany http://www9.cs.fau.de/Persons/Roettger | ||
 |  | Download (14408kb) | 8 bit 0.779724/0.779724/0.791667 FEB03210 | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | ||
 |  | Download (8856kb) | 16 bit 0.693359/0.693359/5 5F420124 | Division of Neuroradiology, University of Erlangen, Germany http://www.neuroradiologie.med.uni-erlangen.de | ||
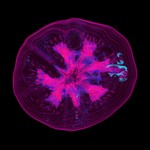 | 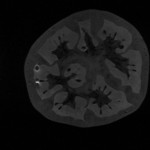 | Download (2292kb) | 8 bit 0.390625/0.390625/1 9872930A | Information and Computing Sciences Division, Lawrence Berkeley Laboratory, USA http://www-itg.lbl.gov | These images and data were produced at Lawrence Berkeley Laboratory by Bill Johnston and Wing Nip of the Information and Computing Sciences Division. This work is supported by the U. S. Department of Energy, Energy Research Division, Office of the Scientific Computing Staff under contract DE-AC03-76SF00098. This data is provided for the purpose of scientific research and collaboration with Lawrence Berkeley Laboratory. Any commercial use of these images and data require prior agreement with Lawrence Berkeley Laboratory. The images and data are Copyright Lawrence Berkeley Laboratory. For further information contact Bill Johnston (510-486-5014, wejohnston@lbl.gov). | |
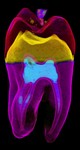 | 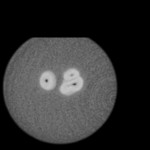 | Download (4540kb) | 16 bit 64E45893 | GE Industrial Micro CT Scanner | From the Transfer Function Bakeoff at IEEE Visualization '00 Conference Original source: http://visual.nlm.nih.gov | |
 |  | Download (7684kb) | 8 bit 0.712691/0.712691/0.796154 AD7982A4 | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | ||
 |  | Download (16964kb) | 8 bit 0.645876/0.645876/0.944063 EDE5D510 | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | ||
 | 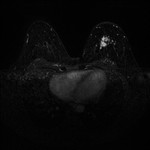 | Download (17072kb) | 16 bit 946A9B34 | Georg-Simon-Ohm University of Applied Sciences, Nuremberg, Germany http://wiki.ohm-hochschule.de/roettger | ||
 |  | Download (11720kb) | RGB 211B2D5B | http://www.virgo.dur.ac.uk | Original scattered data resampled by Matthias Hopf http://www.mshopf.de | |
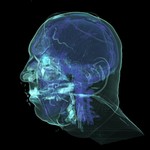 | 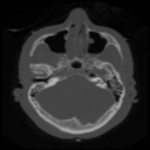 | Download (2792kb) | 8 bit 1.57774/0.995861/1.00797 26B44CE6 | http://www.nlm.nih.gov | ||
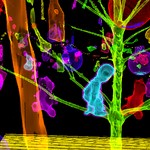 |  | lores version Download (10096kb) | 16 bit 2720D16E | by the Department of Radiology, University of Vienna and the Institute of Computer Graphics and Algorithms, Vienna University of Technology. http://ringlotte.cg.tuwien.ac.at/datasets/XMasTree/XMasTree.html | Real-world dimensions: X-Size: 476mm Y-Size: 476mm Z-Size: 499mm | |
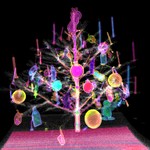 | 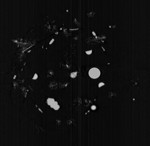 | Download (72236kb) | 16 bit BBE684C1 | by the Department of Radiology, University of Vienna and the Institute of Computer Graphics and Algorithms, Vienna University of Technology. http://ringlotte.cg.tuwien.ac.at/datasets/XMasTree/XMasTree.html | Real-world dimensions: X-Size: 476mm Y-Size: 476mm Z-Size: 499mm | |
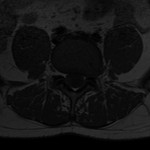 | Download (624kb) | 8 bit 0.661469/0.661469/5.21949 61201BA3 | ||||
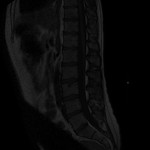 | Download (948kb) | 8 bit 0.682259/0.682259/4.03333 9C968709 | ||||
 | Download (1336kb) | 8 bit 0.392608/0.392608/5.21949 33C61676 | ||||
 | Download (1280kb) | 8 bit 0.546021/0.546021/4.03333 A04FC330 |
This table is generated automatically from my volume data repository
Number of datasets: 85
Last modification: 04.June.2012